gene_interpolation: Spatial interpolation of gene expression
gene_interpolation.RdPerforms spatial interpolation ("kriging") of transformed gene counts
gene_interpolation(
x = NULL,
genes = "top",
top_n = 10,
samples = NULL,
cores = NULL
)Arguments
- x
an STlist with transformed RNA counts
- genes
a vector of gene names or 'top'. If 'top' (default), interpolation of the 10 genes (
top_ndefault) with highest standard deviation in each ST sample is estimated.- top_n
an integer indicating how many top genes to perform interpolation. Default is 10.
- samples
the spatial samples for which interpolations will be performed. If NULL (Default), all samples are interpolated.
- cores
integer indicating the number of cores to use during parallelization. If NULL, the function uses half of the available cores at a maximum. The parallelization uses
parallel::mclapplyand works only in Unix systems.
Value
x a STlist including spatial interpolations.
Details
This function takes an STlist and a vector of gene names and generates spatial
interpolation of gene expression values via "kriging". If genes='top', then
the 10 genes (default) with the highest standard deviation for each ST sample
are interpolated. The resulting interpolations can be visualized via the
STplot_interpolation function
Examples
# Using included melanoma example (Thrane et al.)
# Download example data set from spatialGE_Data
thrane_tmp = tempdir()
unlink(thrane_tmp, recursive=TRUE)
dir.create(thrane_tmp)
lk='https://github.com/FridleyLab/spatialGE_Data/raw/refs/heads/main/melanoma_thrane.zip?download='
download.file(lk, destfile=paste0(thrane_tmp, '/', 'melanoma_thrane.zip'), mode='wb')
zip_tmp = list.files(thrane_tmp, pattern='melanoma_thrane.zip$', full.names=TRUE)
unzip(zipfile=zip_tmp, exdir=thrane_tmp)
# Generate the file paths to be passed to the STlist function
count_files <- list.files(paste0(thrane_tmp, '/melanoma_thrane'),
full.names=TRUE, pattern='counts')
coord_files <- list.files(paste0(thrane_tmp, '/melanoma_thrane'),
full.names=TRUE, pattern='mapping')
clin_file <- list.files(paste0(thrane_tmp, '/melanoma_thrane'),
full.names=TRUE, pattern='clinical')
# Create STlist
library('spatialGE')
melanoma <- STlist(rnacounts=count_files[c(1,2)],
spotcoords=coord_files[c(1,2)],
samples=clin_file) # Only first two samples
#> Warning: Sample ST_mel3_rep1 was not found among the count/coordinate files.
#> Warning: Sample ST_mel4_rep2 was not found among the count/coordinate files.
#> Found matrix data
#> Matching gene expression and coordinate data...
#> Converting counts to sparse matrices
#> Completed STlist!
melanoma <- transform_data(melanoma)
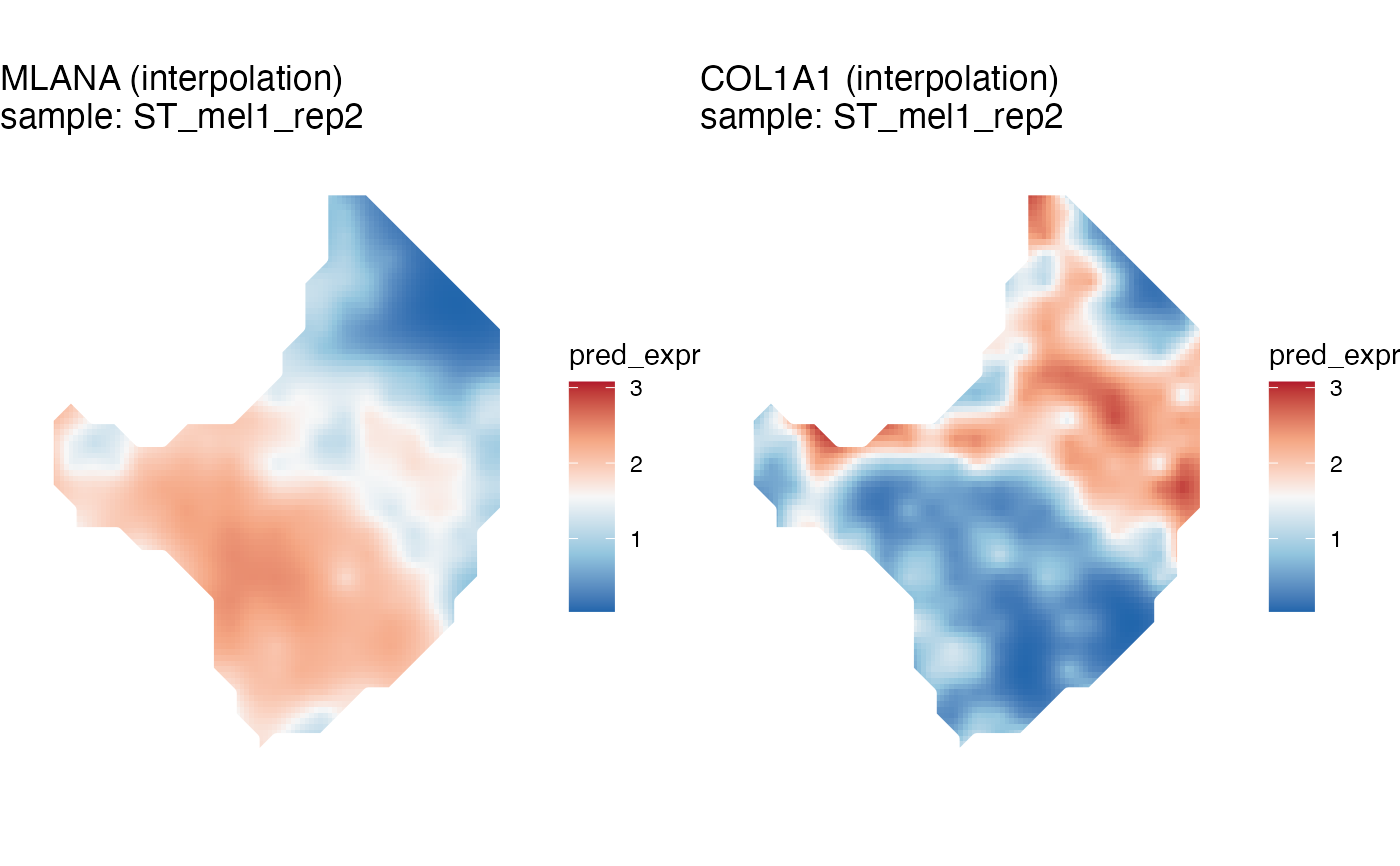
melanoma <- gene_interpolation(melanoma, genes=c('MLANA', 'COL1A1'), samples='ST_mel1_rep2')
#> Gene interpolation started.
#> Warning: No convergence after 200 iterations: try different initial values?
#> [using ordinary kriging]
#> [using ordinary kriging]
#> Gene interpolation completed in 0 min.
kp = STplot_interpolation(melanoma, genes=c('MLANA', 'COL1A1'))
#> Kriging for subject MLANA in sample ST_mel2_rep1 is not present in STlist
#> Kriging for subject COL1A1 in sample ST_mel2_rep1 is not present in STlist
ggpubr::ggarrange(plotlist=kp)